Moreover, we use systems with well-defined compositions to keep the situation as simple as possible and gather fundamental knowledge. Most of our studies focus on the globular proteins myoglobin and lysozyme and the fibrous proteins elastin and collagen. For example, we investigate elastin at low hydration levels to mimic cellular crowding and avoid water freezing in temperature-dependent measurements. Neutron scattering reveals that the mean-square displacement of the elastin protons shows dynamic crossovers at three characteristic temperatures. A comparison with results from nuclear magnetic resonance and broadband dielectric spectroscopy indicates that the crossovers at 125 K and 195 K can be attributed to onsets of methyl group reorientation and protein backbone fluctuations, which occur decoupled from and coupled to water dynamics, respectively, while the crossover at 320 K can be assigned to the calorimetrically detected glass transition.
Institute for Condensed Matter Physics

Dynamics of proteins
The biological functions of proteins require the existence of a hydration shell – or, at least, of an appropriate solvation shell. However, the interplay of protein and solvent dynamics during the biological functions remains elusive to this day.
Crammed with macromolecules
In biological cells, proteins and other macromolecules are densely packed and encompassed only by narrow solvation shells, which comprise water together with various cosolvents. Because of this complexity, we study the relations between protein and solvent dynamics based on appropriate model systems. For example, we use the connective-tissue protein elastin and vary the concentration and composition of the solvent or mimic the crowding in biological cells by confinement to pores. Moreover, temperature-dependent studies allow us to determine the suitability of various cosolvents for the cryopreservation of proteins.
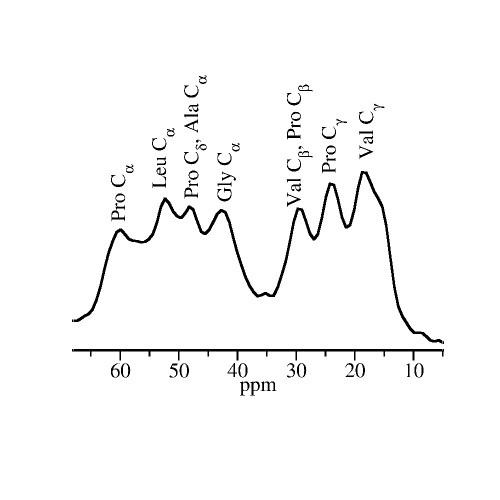
Experiments on protein dynamics
In experimental studies on the relation of protein and solvent dynamics, we combine nuclear magnetic resonance, broadband dielectric spectroscopy, quasielastic neutron scattering, and differential scanning calorimetry to obtain comprehensive insights.
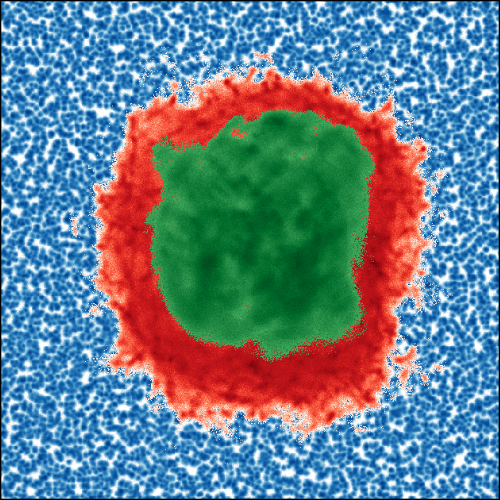
Simulations of protein dynamics
In molecular dynamics simulations, we can fully control the interactions and compositions of the models. Moreover, it is possible to analyze a large variety of properties as a function of space and time. We exploit these capabilities to ascertain the interplay of protein and solvent dynamics.
Selected publications
- Sorin A. Lusceac, Michael R. Vogel, and Claudia R. Herbers (2010). “2H and 13C NMR Studies on the Temperature-Dependent Water and Protein Dynamics in Hydrated Elastin, Myoglobin and Collagen,” Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics 1804, no. 1 (January 2010): 41–48, https://doi.org/10.1016/j.bbapap.2009.06.009.
- Kerstin Kämpf et al. (2020). “Quasielastic Neutron Scattering Studies on Couplings of Protein and Water Dynamics in Hydrated Elastin,” The Journal of Chemical Physics 152, no. 24 (June 28, 2020): 245101, https://doi.org/10.1063/5.0011107.
- Timothy Wohlfromm and Michael Vogel (2019). “On the Coupling of Protein and Water Dynamics in Confinement: Spatially Resolved Molecular Dynamics Simulation Studies,” The Journal of Chemical Physics 150, no. 24 (June 28, 2019): 245101, https://doi.org/10.1063/1.5097777.




